Analysis Of Visium Spatial Transcriptomics Data 2024
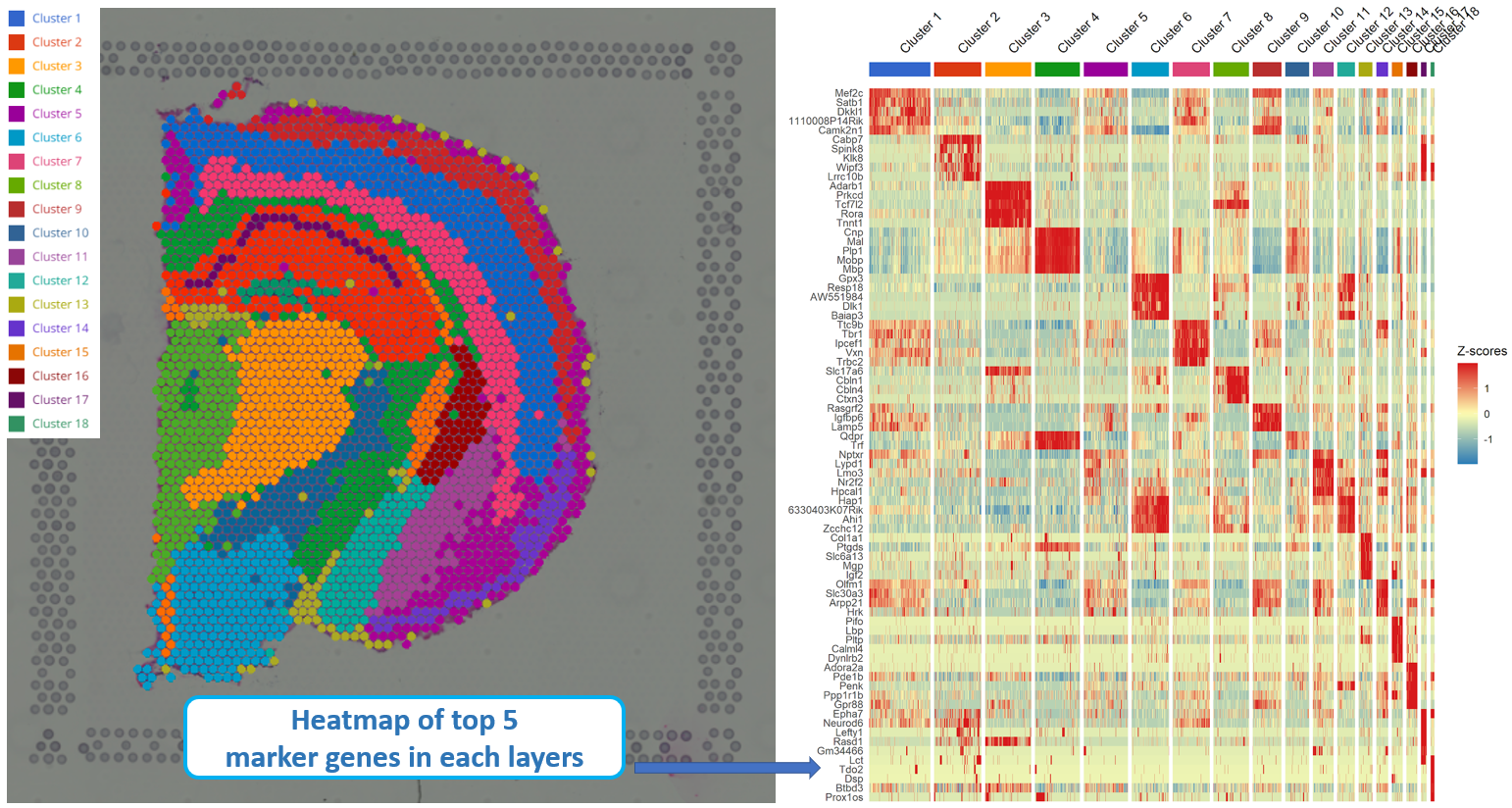
Explore 10x Visium Spatial Transcriptomics Data At Ease With Bioturing Results: here we present spatialone, an end to end pipeline designed to simplify the analysis of 10x visium data by combining multiple state of the art computational methods to segment, deconvolve and quantify spatial information; this approach streamlines the analysis of reproducible spatial data at scale. availability and implementation. Spatial transcriptomic data processing and analysis. raw data for visium assay in fastq files with manually aligned histological images were aligned and quantified using the space ranger software.
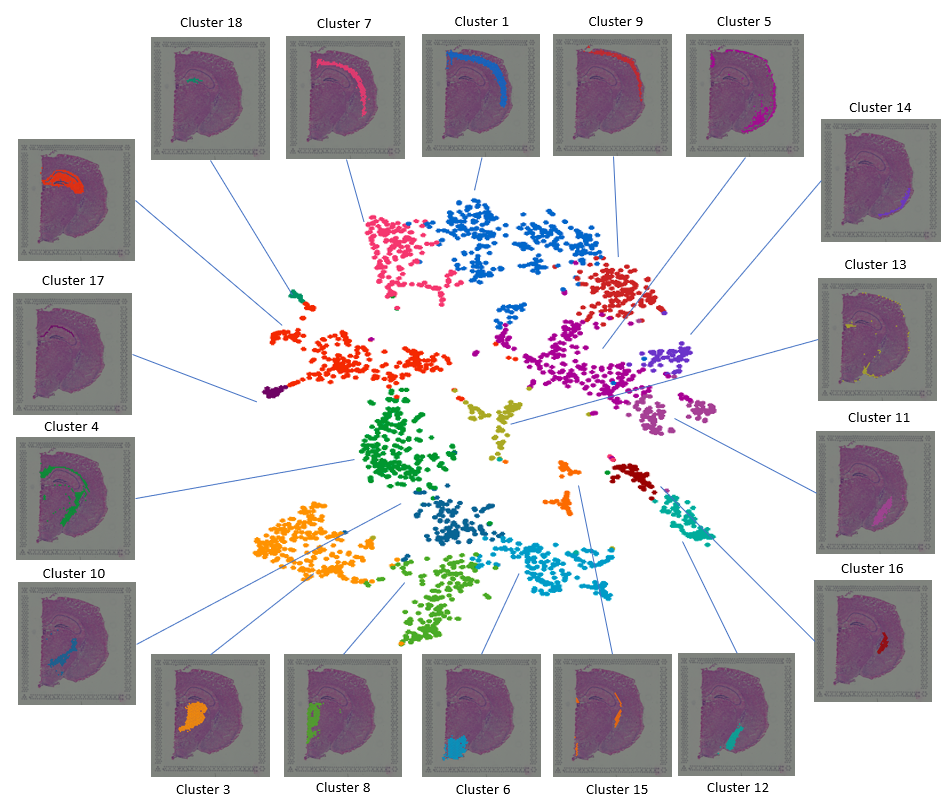
Explore 10x Visium Spatial Transcriptomics Data At Ease With Bioturing Spatial transcriptomics (st) technologies are powerful tools to illustrate the spatial hierarchy and heterogeneity of tissues with the lens of multiplexed gene readouts. however, st technologies generate sequence data rather than images, preventing intuitive examination of the cellular contexture of tissues. moreover, the inherent sparsity of st data caused by molecular crowdedness and. Figure 1. banyan introduces the analysis of community connectivity. spatial transcriptomics platforms yield gene expression and spatial coordinate matrices, which may be used to construct spot spot nearest neighbor networks that describe how cell spots are similar in terms of gene expression or spatial information. Next, we utilized the mouse kidney visium hd dataset to assess redeviz’s enhanced visualization capability at the or gan level. based on the he staining, the kidney can be divided into four layers (figure 4a). after self enhancement, these four layers can be clearly visualized automatically based on the visium hd data (figure 4b). Mil for mapping cccs from srt data. we present a mil model, spacia, to infer cccs from srt data. under a mil framework 12,13,14, the receiver cells are modeled as ‘bags’ with labels.
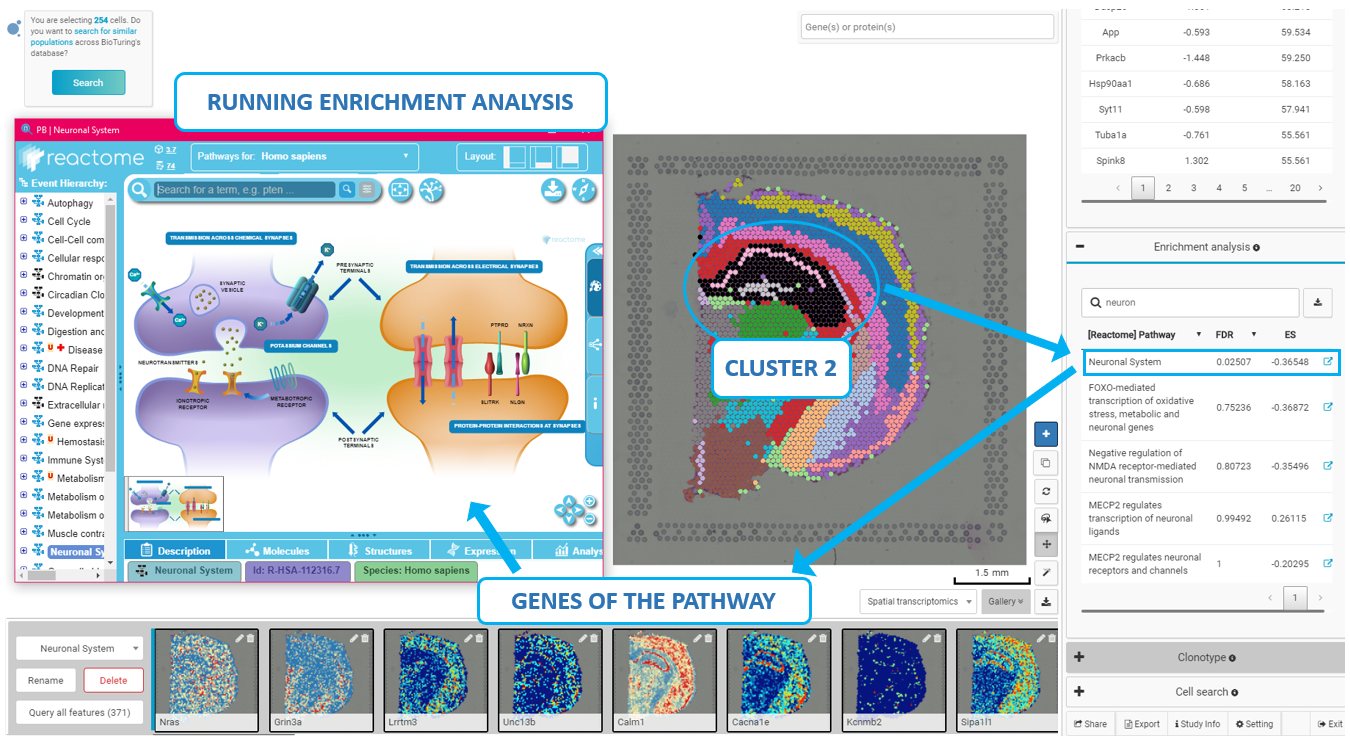
Explore 10x Visium Spatial Transcriptomics Data At Ease With Bioturing Next, we utilized the mouse kidney visium hd dataset to assess redeviz’s enhanced visualization capability at the or gan level. based on the he staining, the kidney can be divided into four layers (figure 4a). after self enhancement, these four layers can be clearly visualized automatically based on the visium hd data (figure 4b). Mil for mapping cccs from srt data. we present a mil model, spacia, to infer cccs from srt data. under a mil framework 12,13,14, the receiver cells are modeled as ‘bags’ with labels. In addition, tumour and background tissue sections from eight patients (of the aforementioned 25) were processed for spatial transcriptomics using the 10x genomics visium platform (n = 36 sections. Existing seurat workflows for clustering, visualization, and downstream analysis have been updated to support both visium and visium hd data. we note that visium hd data is generated from spatially patterned olignocleotides labeled in 2um x 2um bins. however, since the data from this resolution is sparse, adjacent bins are pooled together to.
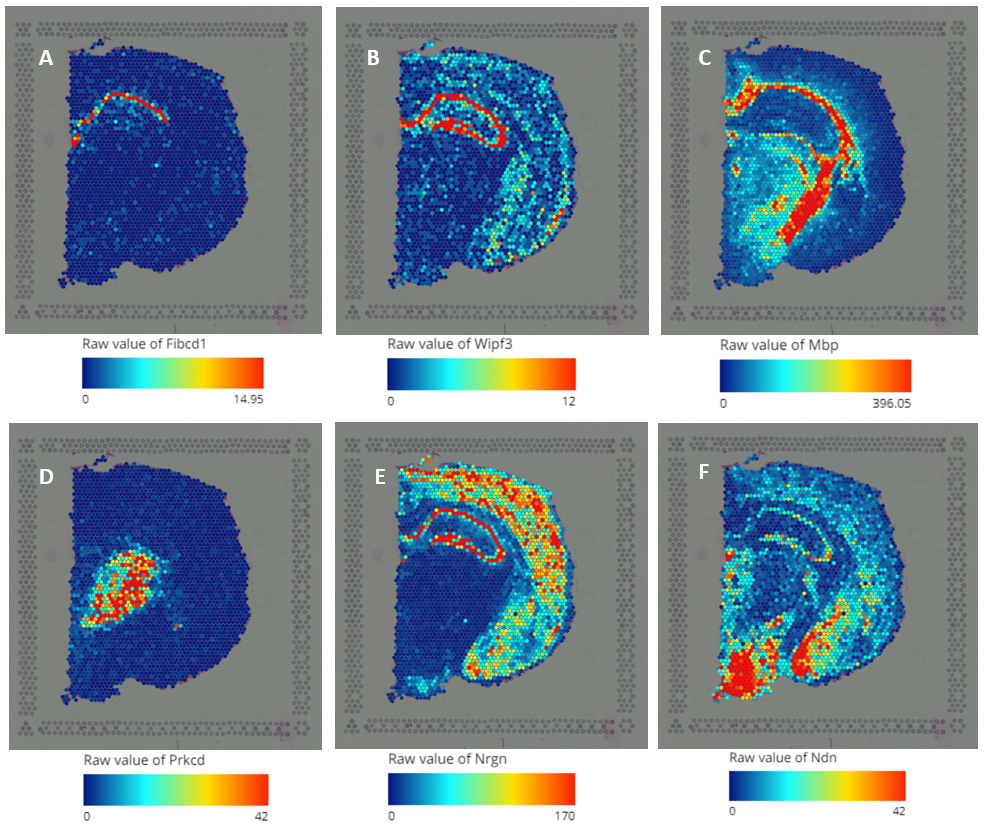
Explore 10x Visium Spatial Transcriptomics Data At Ease With Bioturing In addition, tumour and background tissue sections from eight patients (of the aforementioned 25) were processed for spatial transcriptomics using the 10x genomics visium platform (n = 36 sections. Existing seurat workflows for clustering, visualization, and downstream analysis have been updated to support both visium and visium hd data. we note that visium hd data is generated from spatially patterned olignocleotides labeled in 2um x 2um bins. however, since the data from this resolution is sparse, adjacent bins are pooled together to.
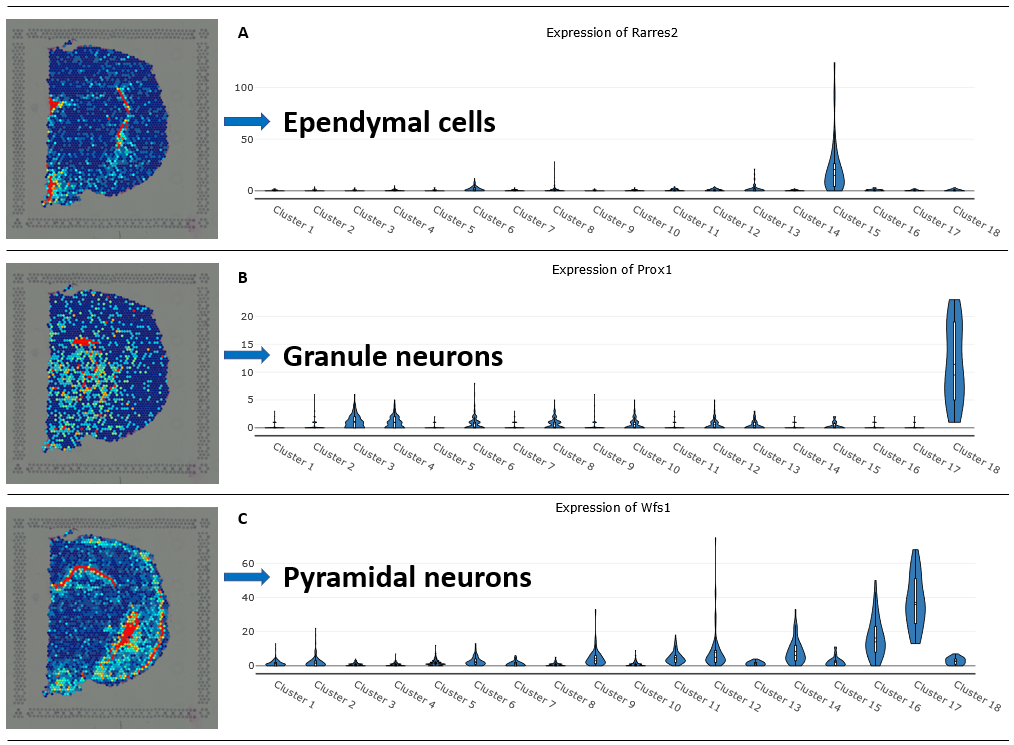
Explore 10x Visium Spatial Transcriptomics Data At Ease With Bioturing

Comments are closed.