Deep Learning Enables Rapid Identification Of Potent Ddr1 Kinase Inhibitors Aisc

Flipboard Genetic Engineering And Human Animal Hybrids How China Is We used six data sets to build the model: (1) a large set of molecules derived from a zinc data set, (2) known ddr1 kinase inhibitors, (3) common kinase inhibitors (positive set), (4) molecules. We used gentrl to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis and other diseases, in 21 days. four compounds were active in biochemical assays, and two were validated in cell based assays. one lead candidate was tested and demonstrated favorable pharmacokinetics in mice.

Deep Learning Enables Rapid Identification Of Potent Ddr1 Zhavoronkov, a. et al. deep learning enables rapid identification of potent ddr1 kinase inhibitors. nat. biotechnol. 37, 1038–1040 (2019) article pubmed google scholar. Gentrl optimizes synthetic feasibility, novelty, and biological activity. we used gentrl to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis and other diseases, in 21 days. four compounds were active in biochemical assays, and two were validated in cell based assays. A machine learning model allows the identification of new small molecule kinase inhibitors in days and is used to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis and other diseases, in 21 days. we have developed a deep generative model, generative tensorial reinforcement learning (gentrl), for de novo small molecule design. gentrl. For slides and more information on the paper, visit aisc.ai.science events 2019 10 23discussion lead: alex zhebrak motivation: we have developed a de.

Deep Learning Enables Rapid Identification Of Mycotoxin Degrading A machine learning model allows the identification of new small molecule kinase inhibitors in days and is used to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis and other diseases, in 21 days. we have developed a deep generative model, generative tensorial reinforcement learning (gentrl), for de novo small molecule design. gentrl. For slides and more information on the paper, visit aisc.ai.science events 2019 10 23discussion lead: alex zhebrak motivation: we have developed a de. Inhibition of ddr1 auto phosphorylation in u2os cells stimulated with collagen. representative blots of phosphorylated ddr1 y513 in u2os cells stimulated with collagen and treated with ddr1 inhibitors at different doses. dasatinib was served as a positive control. dasatinib, compounds 1 and 2 inhibited auto phosphorylation in a dose dependent. Gentrl optimizes synthetic feasibility, novelty, and biological activity. we used gentrl to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis.
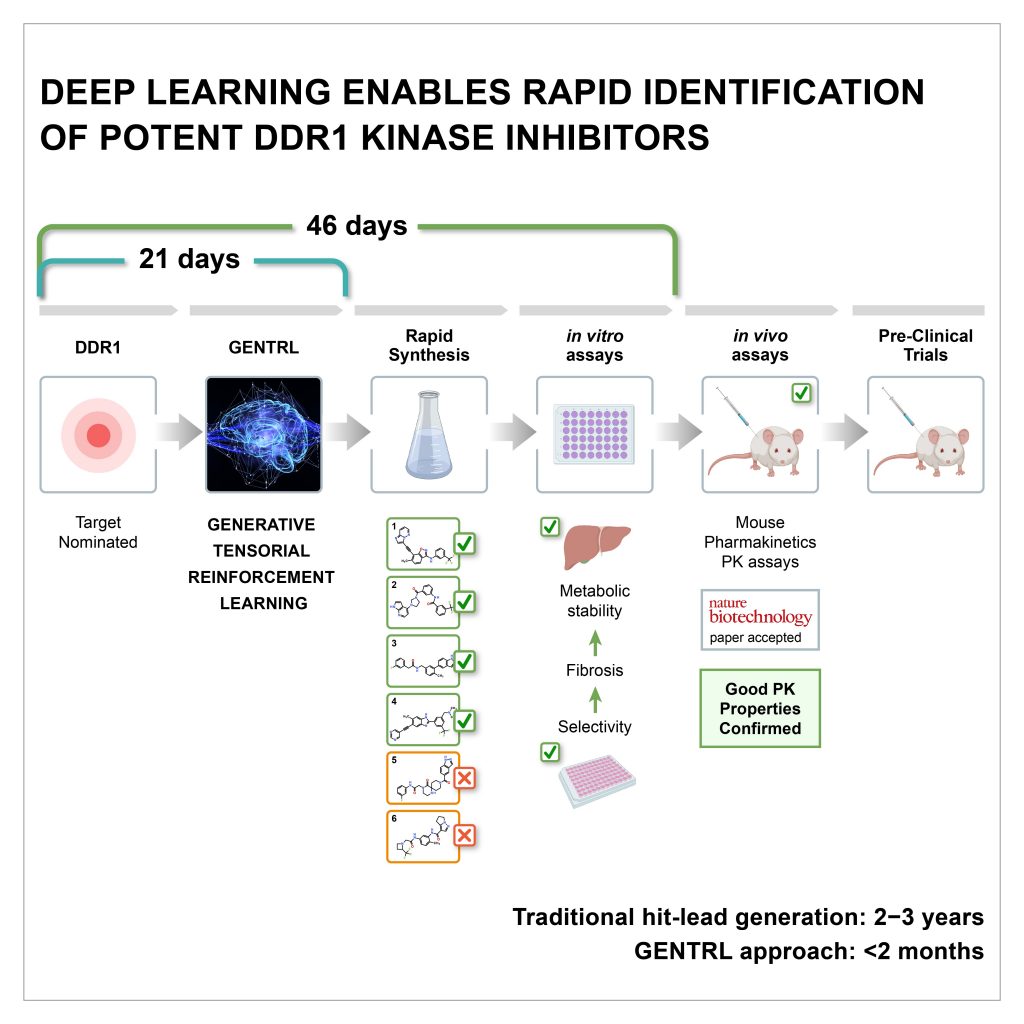
Deep Learning Enables Rapid Identification Of Potent Ddr1 Inhibition of ddr1 auto phosphorylation in u2os cells stimulated with collagen. representative blots of phosphorylated ddr1 y513 in u2os cells stimulated with collagen and treated with ddr1 inhibitors at different doses. dasatinib was served as a positive control. dasatinib, compounds 1 and 2 inhibited auto phosphorylation in a dose dependent. Gentrl optimizes synthetic feasibility, novelty, and biological activity. we used gentrl to discover potent inhibitors of discoidin domain receptor 1 (ddr1), a kinase target implicated in fibrosis.

Comments are closed.