Generation Of Count Matrix Introduction To Single Cell Rna Seq
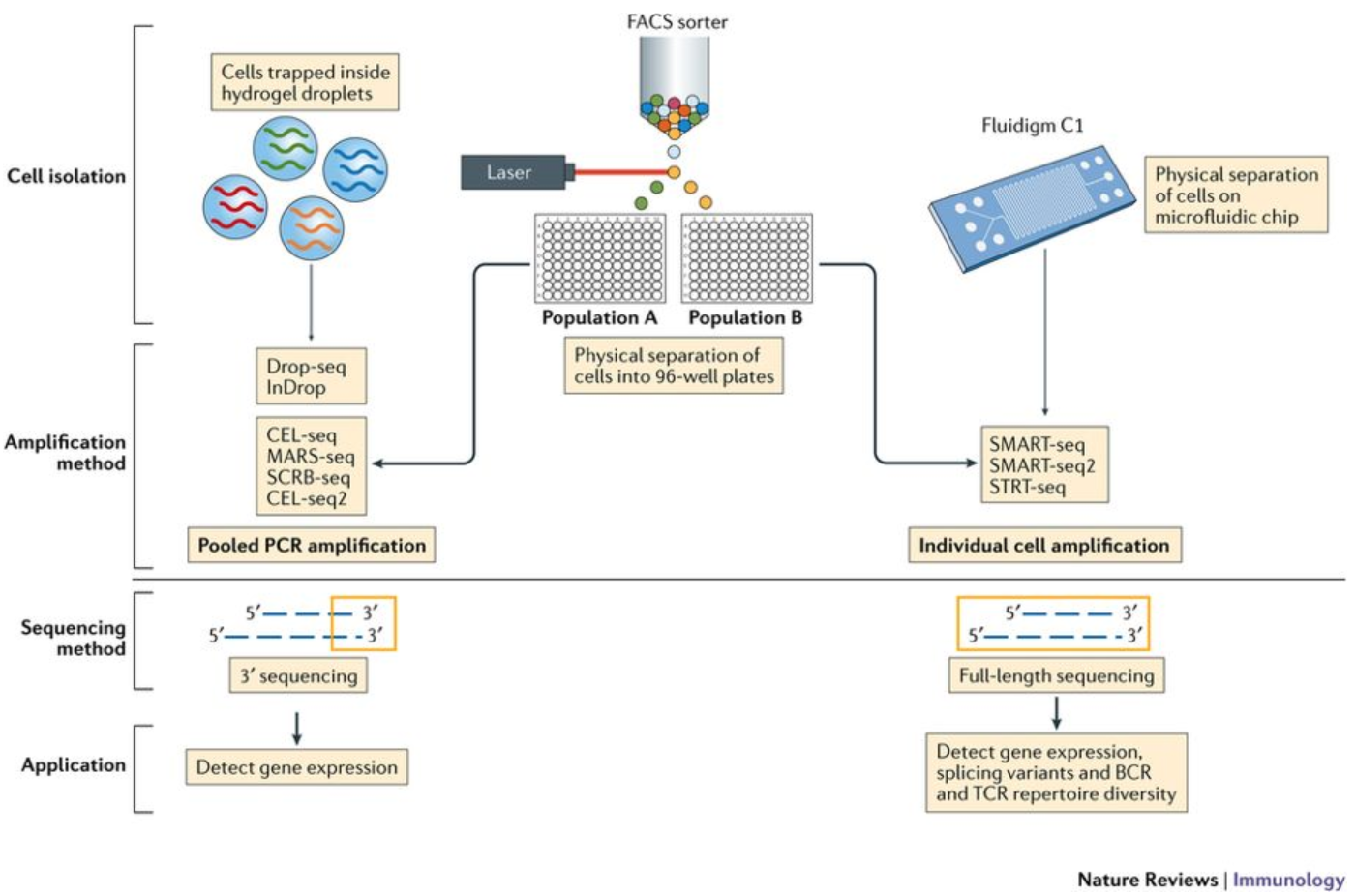
Generation Of Count Matrix Introduction To Single Cell Rna Seq 6696 Skip to the content. generation of count matrix view on github single cell rna seq data raw data to count matrix. depending on the library preparation method used, the rna sequences (also referred to as reads or tags), will be derived either from the 3’ ends (or 5’ ends) of the transcripts (10x genomics, cel seq2, drop seq, indrops) or from full length transcripts (smart seq). The generation of the count matrix from the raw sequencing data will go through similar steps for many of the scrna seq methods. umis and zumis are command line tools that estimate expression of scrna seq data for which the 3’ ends of transcripts were sequenced. both tools incorporate collapsing of umis to correct for amplification bias.

一文了解单细胞 Rna 测序的可视化与统计分析如何更简单高效 知乎 The generation of the count matrix from the raw sequencing data will go through the following steps for many of the scrna seq methods. ’umis provides tools for estimating expression in rna seq data which performs sequencing of end tags of transcript, and incorporate molecular tags to correct for amplification bias.’ the steps in this. 3.1 overview. sequencing data from single cell rna seq experiments must be converted into a matrix of expression values. this is usually a count matrix containing the number of reads mapped to each gene (row) in each cell (column). alternatively, the counts may be that of the number of unique molecular identifiers (umis); these are interpreted. Single cell rna sequencing (scrna seq) is a popular and powerful technology that allows you to profile the whole transcriptome of a large number of individual cells. however, the analysis of the. Seurat is an r package designed for qc, analysis, and exploration of single cell rna seq data. developed and by the satija lab at the new york genome center. it is well maintained and well documented. it has a built in function to read 10x genomics data. it has implemented most of the steps needed in common analyses.
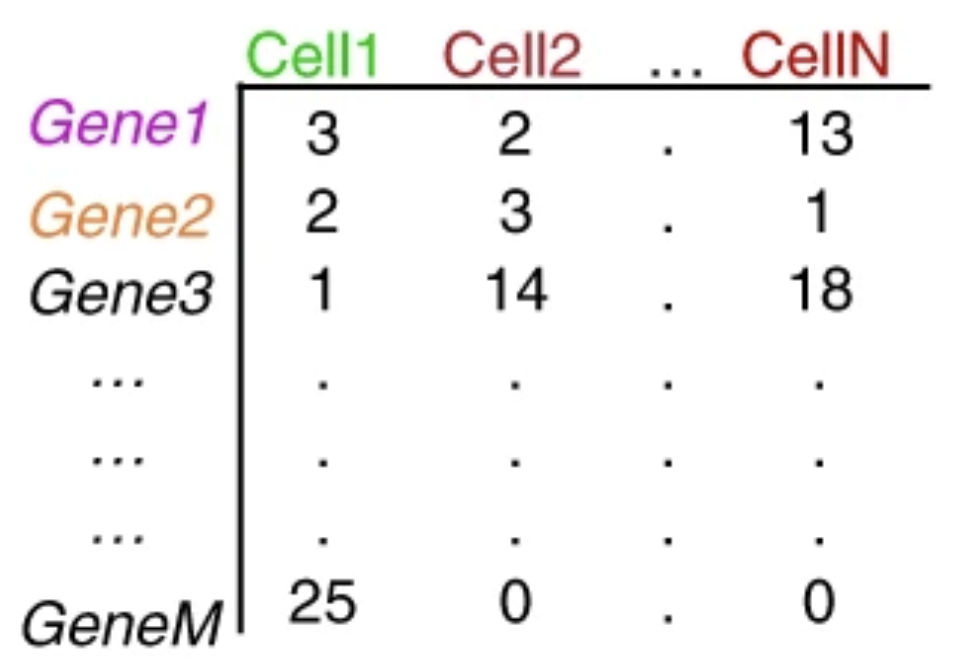
Generation Of Count Matrix Introduction To Single Cell Rna Seq Archived Single cell rna sequencing (scrna seq) is a popular and powerful technology that allows you to profile the whole transcriptome of a large number of individual cells. however, the analysis of the. Seurat is an r package designed for qc, analysis, and exploration of single cell rna seq data. developed and by the satija lab at the new york genome center. it is well maintained and well documented. it has a built in function to read 10x genomics data. it has implemented most of the steps needed in common analyses. Pre‐processing and visualization. raw data generated by sequencing machines are processed to obtain matrices of molecular counts (count matrices) or, alternatively, read counts (read matrices), depending on whether unique molecular identifiers (umis) were incorporated in the single‐cell library construction protocol (see box 1 for an overview of the experimental steps that precede the. A | the first step of single cell rna sequencing (scrna seq) data pre processing involves the generation of a gene expression counts matrix from raw sequencing reads. these reads contain a cdna.

Comments are closed.